v1.5
Credits
['sithu', 'lookang', 'story by jiawen']
Credits
['sithu', 'lookang', 'story by jiawen']
Sample Learning Goals
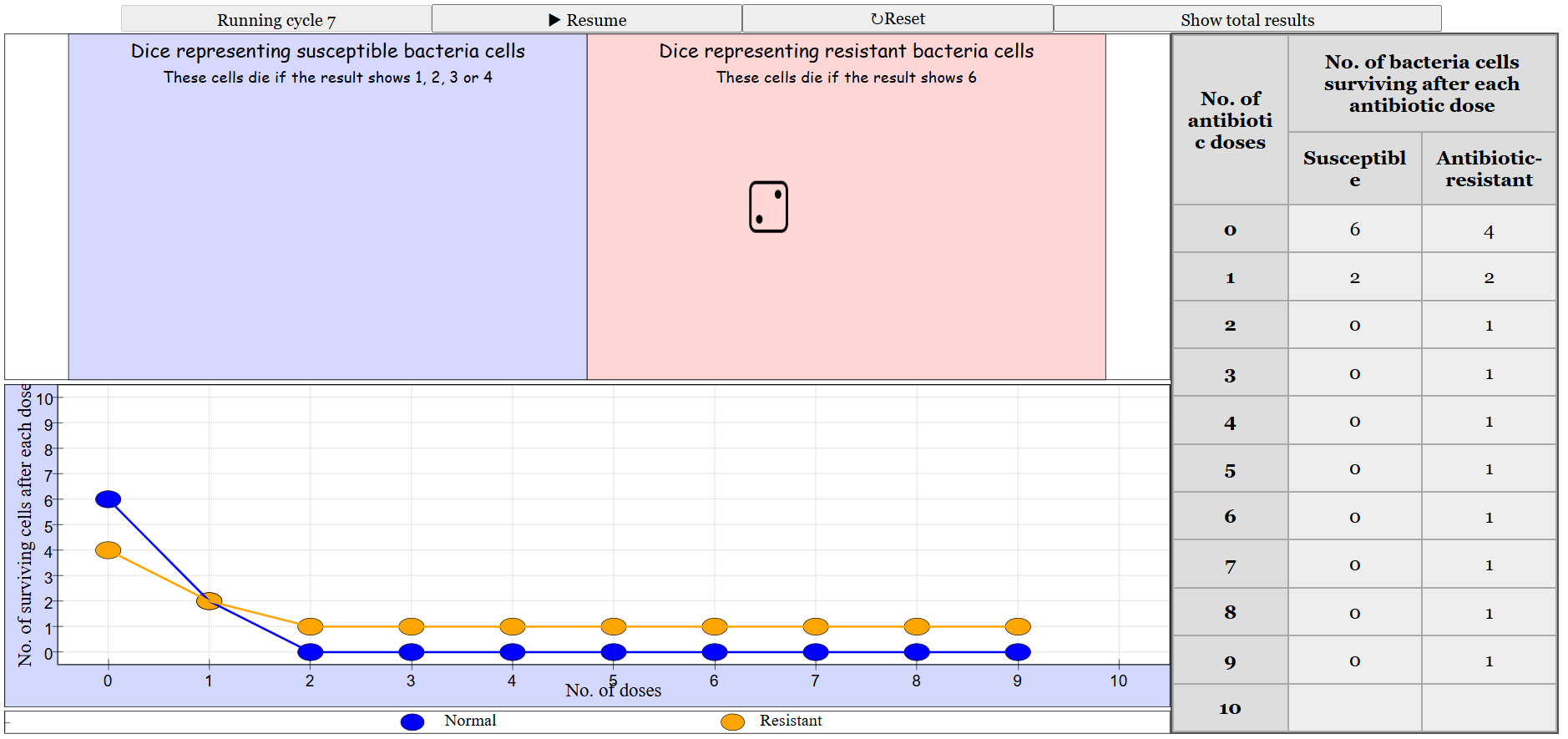
The primary goal of this project is to simulate the effects of antibiotic doses on two populations of bacteria:
- Susceptible (Normal) Bacteria: These bacteria are vulnerable to antibiotics.
- Antibiotic-Resistant Bacteria: These bacteria have developed mechanisms to survive antibiotic treatments.
By modeling these interactions, the simulation aims to educate and illustrate the dynamics of antibiotic resistance, a critical issue in modern medicine.
 |
| link |
Key Features and Functionalities
a. Dual Container Simulation
-
Container A (Susceptible Bacteria):
- Population: Starts with a defined number of susceptible bacteria (e.g., 6).
- Mechanism: Each antibiotic dose affects susceptible bacteria based on a probabilistic model (e.g., rolling a dice where outcomes 1-4 kill a bacterium).
-
Container B (Antibiotic-Resistant Bacteria):
- Population: Starts with a smaller number of resistant bacteria (e.g., 4).
- Mechanism: Resistant bacteria are affected differently (e.g., rolling a dice where outcome 6 kill a bacterium).
b. Simulation Cycles and Doses
- Cycles: The simulation runs through multiple cycles (e.g., 100), each consisting of a set number of antibiotic doses (e.g., 10 per cycle).
- Dose Application: For each dose within a cycle, the simulation rolls dice to determine which bacteria survive or are killed.
c. Death Dose Recording
- Objective: Track and record the specific dose at which all bacteria in each container are eradicated within a cycle.
- Data Recording: Maintains separate records for susceptible and resistant bacteria, indicating the dose number where they were completely wiped out.
d. Interactive Data Display
- Detailed Simulation Table: Shows the number of surviving bacteria after each antibiotic dose for both susceptible and resistant populations.
- Death Dose Record Table: Compiles and displays the dose numbers at which bacteria populations were entirely eliminated across all cycles.
- Toggle Functionality: Users can switch between the detailed simulation view and the death dose records seamlessly during the simulation.
e. Control Mechanisms
- Pause and Resume: Users can pause the simulation at any point and resume it, allowing for interactive exploration and observation.
- Reset Functionality: After completing a set of doses, the simulation resets for the next cycle, maintaining consistent conditions for comparison.
a. Simulation Logic
- Dice Rolling Mechanism: Uses JavaScript intervals and timeouts to simulate the probabilistic killing of bacteria based on dice outcomes.
- Cycle Management: Tracks the number of completed cycles and manages the transition between cycles, ensuring accurate data recording and reset processes.
- Death Dose Tracking: Monitors when bacteria populations reach zero and records the corresponding dose number for each cycle.
b. Data Management
- Arrays for Tracking: Maintains arrays (
normalX,normalY,resistantX,resistantY, etc.) to store data points for each dose and cycle. - Dynamic Table Population: Utilizes DOM manipulation to dynamically add rows to tables, reflecting real-time simulation results.
c. User Interface (UI)
- Responsive Tables: Designed with CSS to ensure tables are scrollable and headers remain fixed, enhancing readability and usability.
- Interactive Controls: Includes buttons or controls for toggling between different data views and managing the simulation state (pause/resume).
Educational Impact
This simulation serves as an educational tool to:
- Demonstrate Antibiotic Resistance: Visually and interactively show how antibiotic-resistant bacteria can survive treatments that eliminate susceptible bacteria.
- Highlight the Importance of Proper Antibiotic Use: Emphasize the consequences of overusing or misusing antibiotics, which can lead to the proliferation of resistant strains.
- Provide Insight into Population Dynamics: Allow users to observe how different factors (e.g., dosage frequency, bacteria populations) influence the overall effectiveness of antibiotic treatments.
For Teachers
[text]
Research
[text]
Video
[text]
Version:
Other Resources
- Details
- Written by Loo Kang Wee
- Parent Category: Interactive Resources
- Category: Biology
- Hits: 2330